Abstract:
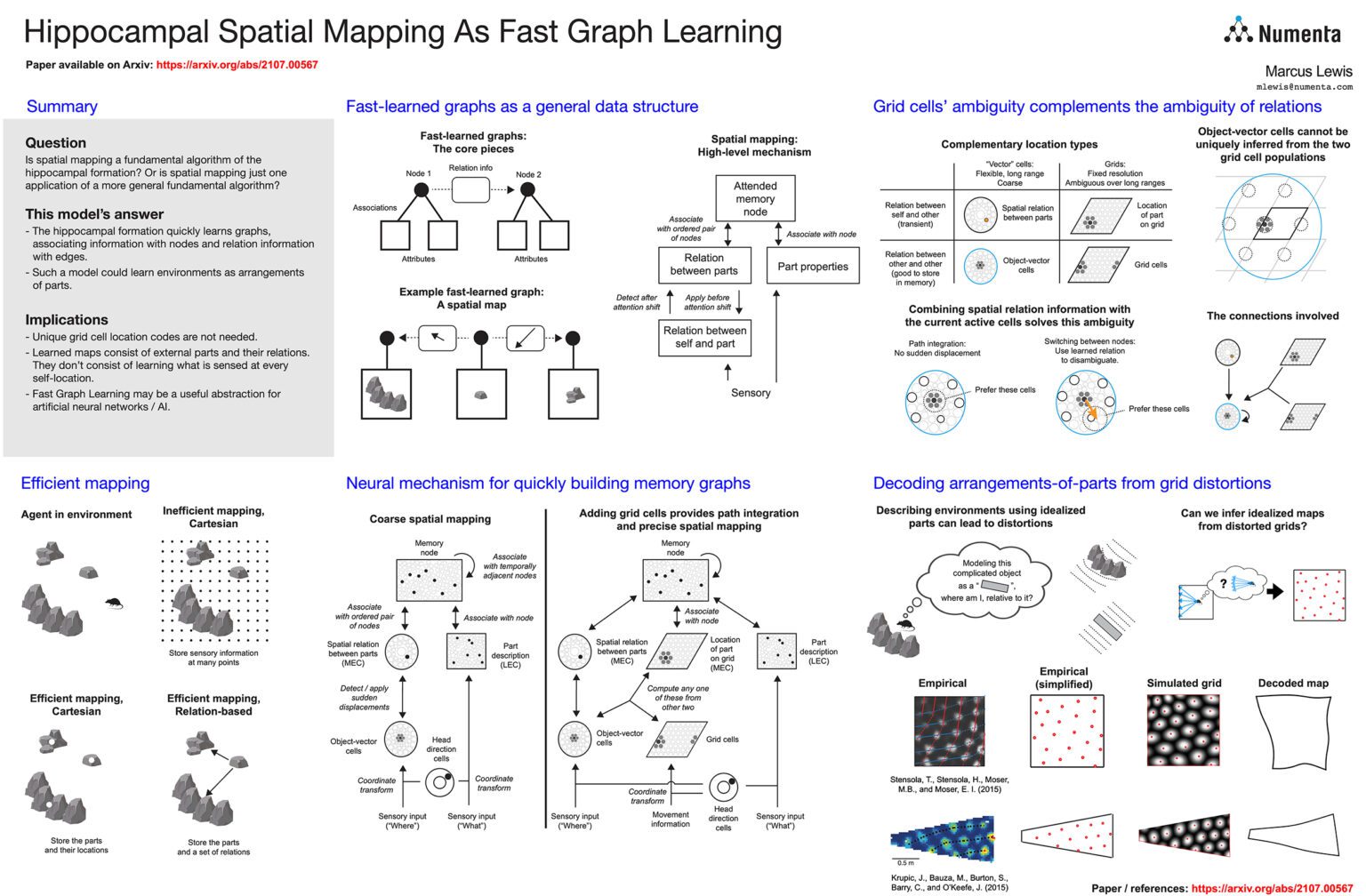
The hippocampal formation is thought to learn spatial maps of environments, and in many models this learning process consists of forming a sensory association for each location in the environment. This is inefficient, akin to learning a large lookup table for each environment. Spatial maps can be learned much more efficiently if the maps instead consist of arrangements of sparse environment parts.In this work, we approach spatial mapping as a problem of learning graphs of environment parts. Each node in the learned graph, represented by hippocampal engram cells, is associated with feature information in lateral entorhinal cortex (LEC) and location information in medial entorhinal cortex (MEC). Each edge in the graph represents the relationship between two parts, and it is associated with coarse displacement information. Thus, the model uses a hybrid approach to storing spatial information, learning ambiguous grid cell locations of environment parts and also learning coarse displacements between those parts. The two complement each other, as the grid cells provide fine-grained resolution that augments the coarse displacements while the coarse displacements disambiguate the grid cells so that a single module is sufficient for unambiguously representing locations.
Using this graph approach, environments can be learned with just a few associations, and the graph can be formed nearly instantly by attending to each of the environment parts. This arrangement-of-parts model offers interesting perspectives on multiple hippocampal phenomena. First, it suggests that each entorhinal module is running an independent mapping system, rather than requiring the modules to work together to represent unambiguous locations. Second, it suggests a reason why grid cells seem to track viewed locations, as that information is exactly what should be associated with nodes in the graph. Third, it offers an explanation for grid cell distortions, suggesting that they occur because the animal fits idealized parts onto actual environment features, and based on this insight we use empirical grid cell data to reconstruct the idealized maps that could lead to such distortions. Fourth, this view explains why hippocampal engram cells are often classified as place cells, suggesting that they actually represent a node in a graph which the animal can attend to from many locations. Fifth, the core idea of associating arbitrary information with nodes and edges is not inherently spatial, so this graph-based view of processing in the hippocampal formation can expand to incorporate non-spatial tasks.
Our model shows that hippocampal modules may dynamically create graphs representing spatial arrangements, and it opens up new ways of understanding how animals make rapid spatial and non-spatial inferences.